

The left-hand image shows RdRp, an enzyme from SARS-CoV-2 that regulates viral replication (stained red), inside a CD4+ T lymphocyte. The right-hand image shows the SARS-CoV-2 spike protein (green) near the cell membrane (image: Henrique Marques-Souza / UNICAMP)
Experiments conducted by Brazilian researchers with lymphocytes isolated from COVID-19 patients showed the virus using the protein CD4 to invade cells that play a key role in coordinating the immune response.
Experiments conducted by Brazilian researchers with lymphocytes isolated from COVID-19 patients showed the virus using the protein CD4 to invade cells that play a key role in coordinating the immune response.
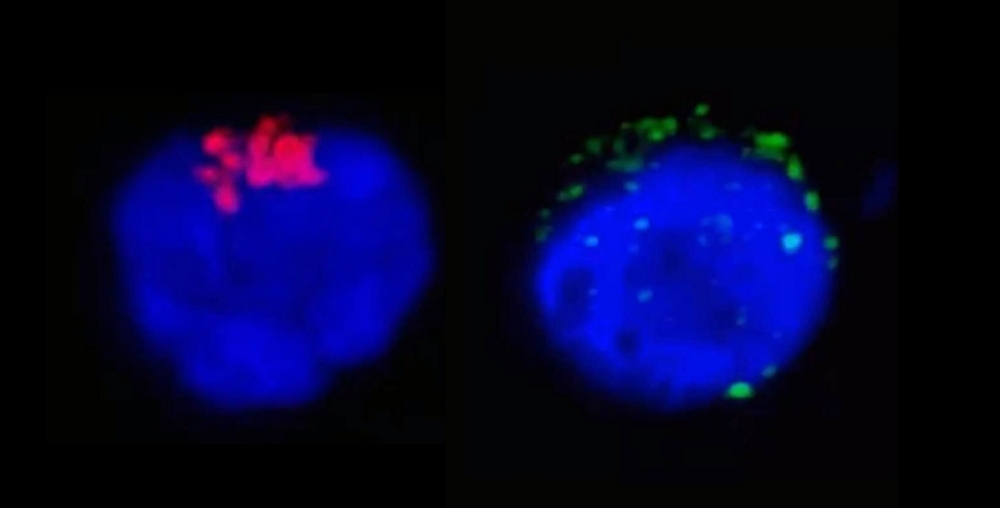
The left-hand image shows RdRp, an enzyme from SARS-CoV-2 that regulates viral replication (stained red), inside a CD4+ T lymphocyte. The right-hand image shows the SARS-CoV-2 spike protein (green) near the cell membrane (image: Henrique Marques-Souza / UNICAMP)
By Karina Toledo | Agência FAPESP – A study coordinated by researchers at the University of Campinas (UNICAMP) in the state of São Paulo, Brazil, and published on medRxiv presents fresh evidence that SARS-CoV-2 can infect and replicate in lymphocytes, potentially inducing the death of these defense cells and at least temporarily compromising the immune system.
According to the authors, the novel coronavirus can be compared to HIV, the virus that causes AIDS, insofar as both affect CD4+ T helper cells, lymphocytes that coordinate the adaptive immune response by having B lymphocytes produce antibodies and helping CD8+ T cells, which recognize and kill infected cells, to proliferate. This coordination occurs via the release of signaling molecules called cytokines.
“Our results suggest that in some patients the novel coronavirus can cause acute immunodeficiency not only by killing some of the subject’s CD4+ T lymphocytes but also by impairing their function. The outcome is that the CD8+ T lymphocytes proliferate less and the B lymphocytes produce antibodies with less affinity and duration. The effect is similar to that of HIV but acute,” Alessandro Farias, co-principal investigator for the study, told Agência FAPESP. Farias heads the Department of Genetics, Evolution, Microbiology and Immunology at UNICAMP’s Biology Institute (IB). The other co-principal investigator was Marcelo Mori, a professor in IB-UNICAMP’s Department of Biochemistry and Tissue Biology.
The findings described in the article, which has yet to be peer-reviewed, were based mainly on experiments with primary cultures of lymphocytes (isolated from the blood of uninfected volunteers and COVID-19 patients) conducted with FAPESP’s support at IB-UNICAMP’s Laboratory of Emerging Virus Studies (LEVE), a Biosafety Level III (BSL-3) facility.
In the first stage of the study, the researchers incubated cells from healthy donors with SARS-CoV-2 and analyzed what happened in the ensuing 24 hours by means of different techniques, such as in situ hybridization, transmission electron microscopy, and RT-PCR (used to diagnose infection in the acute phase).
“We performed this trial only with CD4 and CD8 because they’re the cells that most decrease in severe COVID-19 patients,” Farias said. “The analysis confirmed the presence of the novel coronavirus in some 40% of CD4 cells, with 10% dying by the end of the observation period. The CD8 cells weren’t affected.”
The researchers also found that viral load more than doubled between two hours and 24 hours after the trial began, showing that the virus was replicating in the cultured cells.
Next, they used the same techniques to analyze CD4+ T helper cells isolated from COVID-19 patients in search of signs of SARS-CoV-2. Few of these cells from patients with moderate symptoms were infected. As expected, they produced the cytokine interferon-gamma (IFN-γ), which plays a key role in the immune response. In the case of severe patients, there were many more infected cells, and instead of IFN-γ, they produced interleukin-10 (IL-10), an anti-inflammatory cytokine. In other words, CD4 cells were telling the immune system to stop combating the virus in severe patients.
According to Farias, this would explain why in many severe patients the adaptive immune response (which is specific to each pathogen) displays alterations such as lymphopenia (abnormally low levels of lymphocytes in the blood), T cell depletion, and impaired antibody production.
“Production of IL-10 switches the immune system off and lets the virus stay in the organism for longer,” Farias said. “For now, it’s impossible to distinguish between cause and effect: Did these patients became critically ill because they had more infected CD4 cells or the other way around? In any event, the two factors are clearly associated.”
How the virus gets in
Several scientific papers have shown that SARS-CoV-2 binds to angiotensin-converting enzyme 2 (ACE2) in order to enter human cells, but CD4 cells are known to express a very small amount of this enzyme on the plasma membrane surface, which is coated by the protein CD4 (hence the name of this type of lymphocyte).
To find out how the novel coronavirus gets into CD4 cells, which normally resist infection by viruses and bacteria, the UNICAMP group conducted two more experiments with the blood samples from healthy donors. In the first, antibodies capable of neutralizing CD4 were placed in the cultured cells before they introduced the virus. In the second they used antibodies against ACE2.
“Our hypothesis was that SARS-CoV-2 would succeed in entering the cells using only CD4, but when we also neutralized ACE2 the infection was almost entirely blocked,” Farias said. “This shows that even small amounts of ACE2 are needed for the virus to invade these lymphocytes.”
In vitro molecular interaction tests showed that the SARS-CoV-2’s spike protein can bind to the CD4 in these lymphocytes. “We believe the virus uses a trick to enter these cells. It uses the protein CD4 just to get close to the cell membrane and locate its ACE2 receptor, which is what actually lets it into the intracellular medium,” Farias explained.
In the third and last stage of the study, conducted in partnership with Helder Nakaya, a professor in the University of São Paulo’s School of Pharmaceutical Sciences (FCF-USP), the researchers used bioinformatics to reanalyze data from a study published in May by Chinese scientists in the journal Nature Medicine, reporting the single-cell sequencing of immune cells isolated by bronchoalveolar lavage from the lungs of severe COVID-19 patients.
“The algorithm developed by Nakaya’s group enabled us to identify the viral genome in lymphocytes from the patients’ lungs, providing an additional layer of evidence and enhancing the reliability of the findings,” Farias said.
Researchers at São Paulo State University (UNESP), the Brazilian Center for Research in Energy and Materials (CNPEM), Oswaldo Cruz Foundation (Fiocruz), and D’Or Institute (IDOR) also collaborated. FAPESP supported the study via several grants and scholarships (19/16116-4, 19/06372-3, 20/04583-4, 13/08293-7, 20/04579-7, 15/15626-8, 18/14933-2, 20/04746-0, 19/00098-7, 20/04919-2, 17/01184-9, 19/17007-4, 19/22398-2, 19/05155-9, 19/06459-1, 19/04726-2, 17/23920-9, 16/24163-4 e 16/23328-0).
The researchers are now seeking more detailed knowledge of the effects of SARS-CoV-2’s invasion of CD4 cells with the aim of finding ways to intervene in the process and potentially help combat the infection.
“We already have lymphocytes isolated from more than 350 patients and plan to use them in laboratory experiments, as well as performing tests with mice genetically modified to express human ACE2, among other things to verify the effect of molecules that inhibit the interaction between the viral spike protein and CD4,” Farias said.
The article “SARS-CoV-2 uses CD4 to infect T helper lymphocytes” can be retrieved from: www.medrxiv.org/content/10.1101/2020.09.25.20200329v1.full.pdf.


Republish
The Agency FAPESP licenses news via Creative Commons (CC-BY-NC-ND) so that they can be republished free of charge and in a simple way by other digital or printed vehicles. Agência FAPESP must be credited as the source of the content being republished and the name of the reporter (if any) must be attributed. Using the HMTL button below allows compliance with these rules, detailed in Digital Republishing Policy FAPESP.





